Introduction. The paleogenetic approach becomes an increasingly common component in the archaeological materials analysis and serves as a link between diverse research directions combining it into the one integrated interdisciplinary approach. Currently, the methods of paleogenetics in archaeological and historical studies are very popular and informative. Because they allow to solve the questions of the genetic history of population groups, define their relationships between ancient and modern populations, and do the reconstruction of the sexual, family and social structure of ancient human communities [1-5].
To analyze the populations and individual ethnic groups gene pool and, to determine their basic characteristics, dynamics, history and geography, a huge number of polymorphic markers are used. Among them are high polymorphic loci of coding genes, insertion-deletion polymorphism, micro- and minisatellite or short terminal repeats (STR), single-nucleotide substitution (SNP). From the standpoint of population studies, there are 3 main widely used groups of polymorphic DNA markers: 1) mutations of mitochondrial DNA (mtDNA), transmitted only through the maternal lines; 2) STR and SNP markers of Y-chromosome, inherited only by paternal line; 3) autosomal markers, available for the analysis of maternal and paternal inputs.
The population polymorphism of each group of markers is determined by the factors of microevolution: migration, selection, genetic drift and mutations. In population studies, the variability of a single locus is not significant, but the combination of alleles of closely linked polymorphic loci, which called haplotype, presents a great important.
Polymorphism of mitochondrial DNAs is long ago used in population studies because of the relative simplicity of DNA isolation. This polymorphism type is characterized by the recombination absence and maternal type of inheritance. In this regard, the mitochondrial genome can evolve only through the consistent accumulation of mutations in generations. About 30% of mtDNA variability determines the interpopulation and intergroup differences.
The great potentialities of the Y-chromosome, as a tool for evolutionary research, are associated with its haploidity. Y-chromosome is transmitted strictly along the paternal line and is not involved in recombination processes, in which other chromosomes exchange by alleles of different historical origin. More than 95% of the Y-chromosome length is represented by a non-recombining site. Y-chromosome undergoes only spontaneous mutational changes, and it transmits from generation to generation in a more permanent form than other genome parts, specifically as a separate haplotype represented by a combination of a large number of polymorphic alleles. 59% of Y-chromosomes genetic diversity is due to interindividual differences, 25% references to interpopulation differences, and 16% - to diversity between geographic regions [6, 7]. Another important population characteristic of the Y-chromosome is the very high degree of geographical differentiation, which is much higher than in other genome regions. Based on these properties, Y-chromosome polymorphism is estimated as a sensitive system for determining population migrations within the large regions and continents.
To analyze the origin of ethnic groups and establishing the links between ancient and modern populations, the large-scale studies of archaeological finds and modern populations based on wide genome sequencing are gaining the great importance because they can provide the information about various types of polymorphism [1-5].
In the frame of project entitled “Ancient treasures of East-Kazakhstan”, the massive archeological studies are carried out in East-Kazakhstan region by the groups of Z.Samashev and other leading archaeologists. Among them there are famous necropolis Berel in Katon-Karagai district, Shilikty in Zaisan district, Ablaikit in Ulan district, Kyrykungir in Abai district, and necropolis Eleke-Sazy in Tarbagatai district.

Figure 1 – Golden Man from necropolis Eleke-Sazy
The most important find of 2018 (archaeologist Z. Samashev) was so called Golden Man from Eleke-Sazy pantheon (figure 1). A well-preserved skeleton of a young man of 17–18 years old (height is approximately 165–170 cm) of a Europeoid physical type in a rich military equipment was found in the burial (mound 4). Warrior undoubtedly belonged to the elite of Saka society. This is evidenced by the nature of the burial, a large number of gold items of clothing and military equipment, as well as traces of posthumous mummification of the object. Together with Golden Man, a woman of 11-13 years old was buried in mound 4. In the mound 8, the bone remains of an individual of an anthropologically unidentified sex were found, characterized by age as 35-40 years old. The artifacts were dated by the Early Saka period (VIII-VII centuries BC).
The aim of this study was paleogenetic analysis of paternal and maternal lines of 3 objects representing elite Sakas from Eleke-Sazy necropolis.
Materials and methods.
Study objects. The objects of this paleogenetic study were human bone remains (teeth and samples of Glenoid Fossa from temporal bone (Cochlea)) of 3 individuals from the mounds 4 and 8 of the Eleke Sazy necropolis (Tarbagatai district, East Kazakhstan region).
The sampling (figure 2) was done on September, 7th, 2018 in the on the territory of the Regional Museum of History and Local Lore, Administration of Culture, Archives and Documentation of the East Kazakhstan region. The object Golden Man was presented in the exhibition hall. Another objects from the mounds 4 and 8 were provided by anthropologist Egor Kitov.
Selecting bone fragments, whole teeth were taken (4) and with the help of a Dremel 4000 drill a fragment of Glenoid Fossa from Petrous bone was cut out on the one side of each object's skull. During the sampling of bone remains, to avoid unnecessary contamination of the objects by the foreign DNAs, we tried to observe the principles of sterility, which were available for the current conditions: treatment of surfaces by the 70% ethanol, work in lab coats, gloves and masks, sterile packaging of research materials. Before treatment, bone materials were measured using a metric system.

Figure 2 – Sampling of bone remains of Eleke-Sazy individuals
Pretreatment of bone sampleas and paleo-DNA extraction. Before preparing the bone powder, mechanical cleaning of bone fragment surfaces was performed using emery discs of Dremel 4000. The teeth were first divided in half transversely, coronary dentin was cut out with the help of a dental drill. Then followed the consistant washing of bone fragments by deionized water and 96% ethanol. After drying, to destroy the foreign DNAs on the surface layers, the bone fragments and teeth were packed in labeled sterile packets, and UV-treatment for 15 minutes (each side) was carried out.
Further, all procedures were performed under very sterile conditions in specially equipped rooms (clean area). The bone fragments were ground to prepare bone powder. Grinding was performed using a drill and a TissueLyser II homogenizer (Qiagen, Germany) in the 30 Hz mode, for 40 seconds.
50 mg of bone powder was taken to isolate the paleo-DNAs. Decalcification was performed using 0.5 M EDTA (pH 8.0) at 37°C on a shaker at a speed of 1000 rpm for 16 hours.
Paleo-DNA extraction was performed according to the protocol of Dabney et al. [8] with minor modifications. 2 types of control were used: positive (DNA of an ancient bear from the Pleistocene cave) and negative (only reagents without a sample).
Extraction of ancient DNA was performed using lysis buffer (100 µM EDTA, 0.25 mg/ml proteinase K), incubating overnight (16 hours) at 37°C. Next, bone powder incubated with extraction buffer was precipitated by centrifugation at 2000 rpm for 15 minutes. The collected supernatant was added to 10 ml of binding buffer (5M GuHC1, 40% Isopropanol, 100 mM NaAc). This containing DNA solution was transferred onto silica membranes in MinElute Columns (Qiagen, Germany) columns in the presence of the hemotrophic guanidine hydrochloride salt. The solution was centrifuged at 12,000 rpm for 2 minutes. The bound DNA membranes were washed with 2 times 720 μl of a commercial PE wash buffer (Qiagen, Germany). After drying, DNA elution was performed using 100 μl TET buffer (1 nM EDTA, 10mM Tris-HC1, 0.05% Tween-20).
Quantitative and qualitative evaluation of DNA samples and paleo-DNA degradation was evaluated using a commercial reagent kit on a 2100 bioanalyzer (Agilent Technologies, USA) according to the manufacturer's protocol.
TagMan analysis. Genotyping of Y-chromosome haplogroup specific SNPs (ISOGG 2018, version 13.307 from 24.12.2018) was done by the RT-PCR using the TaqMan probes (ThermoScientific, USA) according to the manufacture protocols. TaqMan probes were aimed for the 26 SNPs: M17, M20, M184, M207, M217, M231, M215, M242, M285, M253, M306, M343, M324, M417, M449, M479, M511, M514, M516, P15, L415, L460, L581, CTS5368/Z2215, PF4610, L901.
Library preparation and NGS analysis. Library preparation for NGS was done according to standard protocol [8]. 20 μl DNA extract was subjected to partial uracil-DNA-glycosylase (UDG) treatment. To obtain blunt ends, the extract was combined with 30 μl of the reaction mixture (1x NEB buffer 2, 100 μM dNTP mix, 0.8mg/ml BSA, 1mM ATP, 0.4U/μl T4 Polynucleotide Kinase (Biolabs, UK), 0.024U/μl T4 Polymerase, (Biolabs, UK), then incubated for 15 min at 15°C and for 15 min at 25°C. The same treatment was carried out in the case of negative and positive control. DNA was purified using MinElute column (Qiagen, Germany). 18 µl of DNA solution was used for ligation of adapters in reaction mixture containing 1xQuick Ligase buffer, 250 nM Solexa Adapter Mix and 0.25U/1 Quick ligase (Biolabs, UK), which incubated for 20 min at room temperature. Then the MinElute column (Qiagen, Germany) were used again for DNA purification. 20 µl of DNA was used for the “filling reaction” in reaction mixture containing 1xIsothermal buffer, 125Nm dNTP, 0.4U/µl Bst polymerase (Biolabs, UK), that was incubated for 20 minutes at 37°C, then - for 20 min at 80°C without further purification. DNA library fragments were stored at -20°C. was used for the quantification of library fragments was done using the DyNAmo HS SYBER Green Q PCR Kit (ThermoScientific, USA), IS7 and IS8 primers according to the manufacture’s protocol. The dilutions of standard from 1 to 10 copies/µl were performed. PCR conditions were 95°C 10 min, 60°C 30 sec, 72°C 30 sec.
50 ng preamplified libraries were double indexed as described by Kircher M. et al. (2012) [10] in a 100 μl PCR reaction mixture containing 0.25U/µl Pfu Trubo Polymerase (Agilent Technology, USA) and 1000 nM each of IS5 and IS6 indexing primers. PCR conditions were 95°C 2 min, 6 cycles of 58°C 30 sec, 72°C 1 min followed by a final extension of 72°C 10 min. Indexed libraries were MinElute purified and their concentration was measured with Qubit, and size distribution was checked on Agilent 2200 TapeStation Genomic DNA ScreenTape.
Biotinilated mtDNA baits were prepared from three overlapping long-range PCR products according to [11], but using the primers described in [12]. Capture was done according to [11]. PCR conditions were: 98°C 1 min, 10 cycles of 98°C 20 sec, 60°C 30 sec, 72°C 30 sec, followed by a final extension of 72°C 30 sec. The captured and amplified library mix was purified on MinElute column and eluted in 15 μl EB. Before sequencing, libraries were quantified with Qubit, and quality checked and Agilent 2200 TapeStation Genomic DNA ScreenTape. Sequencing was done using MiSeq analyzer (Illumina, USA) with MiSeq Reagent Kit v3 (MS-102-3003) generating 2x150 bp paired-end sequences.
Data analysis. DNA sequencing data were processed using the EAGER v1.92.50 pipeline [13]. From VCF files FASTA format was generated with the Genom Analysis Tool Kit (GATK v3.5) Fasta Alternate Reference Maker walker. Sequences shorter than 25 nucleotide were removed from this dataset. Mitochondrial haplogroups were determined using both HaploGrep2 and HaploFind for Mt-calls. The Y haplogroup was determined using the yHaplo program.
Results and their discussion. Paleo-DNAs extracted from ancient bone fragments and teeth taken from Eleke-Sazy necropolis allowed to create suitable libraries for NGS. Quantative and qualitive charachteristics of DNA-fragments of libraries represents table 1.
Table 1 – Charachteristics of paleo-DNAs libraries
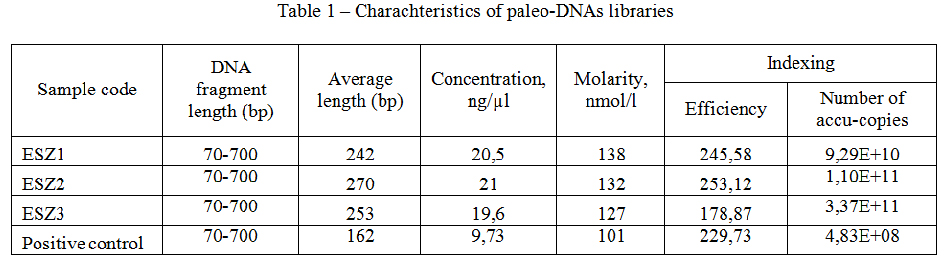
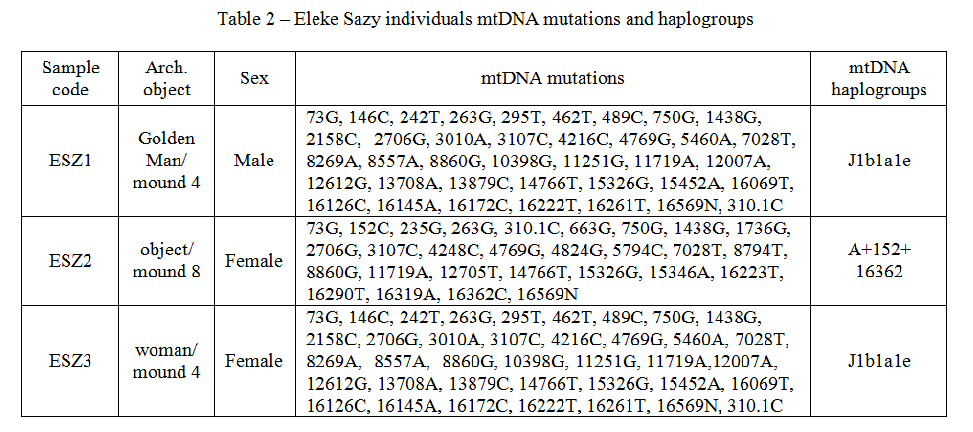
It was established, that anthropologicaly undefined individual from Eleke sazy mound 8 was a woman carrying A haplotype of mtDNA. Golden Man and young woman buried in the mound 4 had the same J1b1a1e mtDNA haplotype, that indicates the relationship from maternal side of both individuals. Ansient elite Saka objects mitochondrial DNA mutations and haplogroups are represented in table 2.We have successfully sequenced all 3 ancient objects DNAs. The number of sequenced reads was quite efficient for ancient samples: ESZ1 (Golden Man/mound 4) - 4526052; ESZ2 (object/mound 8) - 49954656; ESZ3 (woman/mound 4) – 24395572. Contamination of mtDNA was estimated as 0,00 (for ESZ2) and 0,01 (for ESZ1 and ESZ2). Estimation of nuclear DNA contamination of revealed following: ESZ1 - 0,0028; ESZ2 - 0,1967; ESZ3 – 0,2611.
Table 2 – Eleke Sazy individuals mtDNA mutations and haplogroups
According to our data, representatives of the J (J, J1d1a, J1c2, J1c16, J1d, J2b1c1, JT) haplogroup mtDNA in the cohort of studied modern Kazakhs were met with a total frequency of 3,7% (9/246), without tribal specificity.The J mtDNA clade derives around 45,000 years before present. Based on ancient DNA tests, haplogroup J firstly met at woman who lived in the Near East or Caucasus. The time of J1b subclade appearance were estimated as 23,300 years (± 4,300 years) in Near East and as 5,000 years (± 2,200 years) in Europe. J1b1a found in Europe, the Caucasus and India, J1b1a1 found in western Europe. There were no data about J1b1a1e subclade origin.
Haplogroup J averages 7,6% and varies from 4,4% among Georgians to 10,9% in Poles. Díaz-Zabala H.J. with coauthors [14] noted that in the Puerto Rican population haplogroup J distributed with a frequency of 23% and the most frequent subclade of this haplogroup is J1b1a1 (16%). Ancient DNA studies have associated J1b1a1 with the diffusion of Proto-Germanic and Proto-Celto-Italic speakers, because its frequencies are higher in North-Central Europe. However, there is an opinion that some populatioons such as Moroccan Jews, or Turks could be responsible for the distribution of J1b1a1 in Europe. Battles between the Ottoman Empire and Spanish-led Catholic coalitions were common during the 16th century, and could have led to the introduction of Near Eastern mtDNAs to Spanish colonies in the Americas [14].
We suppose that distribution of J mtDNA haplogroups and their subclades was related with migrations of Paleolithic populations to central part of Eurasia from Near East through Caucasus. Possibly the carriers of J1 mtDNA clade migrate to the Central Eurasian Steppe and Altai mountains regions before Early Bronze Age. The quite high frequencies of J1 and related subclades (J1a, J1c, J1d and others) in Central Asia are evidenced this hypothesis. And 146C mutation of mtDNA HVR2 region that related with J1b1a1e subclade could be appeared in early Saka tribes of Altai.
Haplogroup A of mtDNA is more older than J1b1a1e subclade. The appearance of A haplogroup is believed took place in Asia about 30,000–50,000 years before present. Quite low diversity of A haplogroups subclades supports the opinion that it could be descended from a population that has emerged from a bottleneck approximately 20,000 years ago.High distribution of A haplogroup and related subclades between ancient and modern populations of China, Mongolia, Japanese, Tibet, Altai, Siberia, Far East and very rare frequencies in West Europe supports the idea that it might have originated in and spread from the Far East. The A highest frequencies are among Native Americans, its largest overall population is in East Asia, and its greatest variety is in East Asia. Our data represent the variety of A subclades (A, A5a, A14, A15, A16, A+152+16362) at modern Kazakhs - 5,69% (14/246). It should be noted that 50% of mtDNA A haplogroup carriers belong to Kazakh tribe Kerey (Middle zhuz), which related to East Kazakhstan region.But 2 individs belonging to Junior zhuz (Alimuly and Baiuly tribes) and 1 individ representing the Senior zhuz (Suan tribe) have the same maternal line (mtDNA haplotype A+152+16362) with ancient woman from Eleke-sazy mound 8.
The analysis of Y-chromosome SNPs has revealed that paternal line of Saka Golden Man belongs to R1a1a1-M417 haplogroup. Table 3 represents Y-chromosome haplogroup-specific SNPs identified at paleo-DNA of Eleke Sazy ancient man.
Table 3 – Y-chromosome SNP-markers identified at Eleke Sazy ancient man
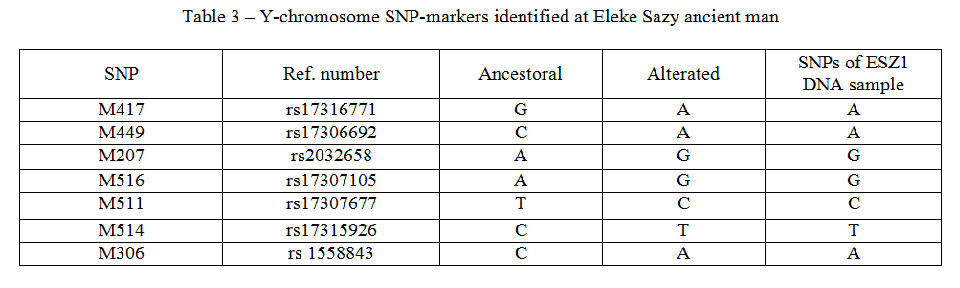
According to the Underhill P.А. with coauthors [15] the R1a1a1 defining SNPs are М207, М173, М306, М420, М449, М511, М513, SRY10831.2, М515, М448, М459, М516, М17, М198, М512, М514, М515, and М417. The 7 SNPs from this list were identified at elite Saka from Eleke Sazy necropolis. All identified SNPs correspond to the analyzed subclade of R1a.
While R1a originated about 25,000 years ago, its subclade M417 (R1a1a1) diversified ca. 5,800 years ago [15]. The authors believe that diversification occurred in the vicinity of Iran and Eastern Turkey. This suggests the possibility that R1a lineages accompanied expansions of ancient populations initiated during the Copper, Bronze, and Iron ages, partially replacing previous Y-chromosome strata, an interpretation consistent with limited ancient DNA evidence [12]. The distribution of M417-subclades in Central and Eastern Europe and in Asia suggests that R1a1a diversified within the Eurasian Steppes between Middle East and Caucasus. The place of origin of thise subclade plays a role in the debate about the origins of Proto-Indo-Europeans [15].
R1a1a1 (R-M417) is the most widely found subclade, in two variations which are found respectively modern population of Europe (R1a1a1b1-Z282) and Central and South Asia (R1a1a1b2-Z93). Our data show that the total frequency of R1a and R1a1a-M17 at modern Kazakhs is 9.58% (166/1732). R1a-M17 and R1a1a-M17 were found with low frequencies among representatives of many different tribes. The most representative imput of these clades were noted in the representatives of Argun-Momyn-Atygai (57.69%), Zhetiru-Tabyn (32.0%) and Argun-Momyn-Tobykty (22.72%) tribes. This circumstance testifies the fact that R1a subclade of Y-chromosome was quit popular among paternal lines of Saka populations distributed throughout the vast territory of present Kazakhstan in Early Iron age.
We hope that the whole genome analysis of Eleke Sazy individuals and other Elite Saka from the Kazakhstan territory, which is doing in collaboration with Johannes Krause team (Department of Archaeogenetics, Max Planck Institute for the Science of Human History, Jena, Germany), will provide more detailed information not only about the origin of the paternal and maternal lines of the Elite Sakas from Eleke Sazy necropolis, but also determine the migration history of this ancient population, and define the phylogenetic relationships between the associated Eurasian Saka communities.
Acknowledgement: This work was supported by the Akimat of the East Kazakhstan Region for 2018-2019 and on research work on the topic: “The history and culture of the Great Steppe” (BR05233709). The authors are also grateful for the joint analysis of the Johannes Krause team (Department of Archeogenetics, Max Planck Institute for the Science of Human History, Jena, Germany).
REFERENCES
1 Haak W., Lazaridis I., Patterson N. et al. Massive migration from the steppe was a source for Indo-European languages in Europe // Nature. – 2015. – Vol.522(7555). – P.207-211. doi: 10.1038/nature14317.
2 Krause J. and Pääbo S. Genetic time travel // Genetics. – 2016. – V.203. – P.9-12.
3 Siska V., Jones E.R., Jeon S. et al. Genome-wide data from two early Neolithic East Asian individuals dating to 7700 years ago // Science Advances. – 2017. – Vol.3(2). doi: 10.1126/sciadv.1601877
4 Jeong C., Wilkin S., Amgalantugs T. et al. Bronze Age population dynamics and the rise of dairy pastoralism on the eastern Eurasian steppe // Proc. Natl. Acad. Sci. – 2018. – Vol.115(48). – P.11248-11255. doi: 10.1073/pnas.1813608115.
5 Narasimhan V.M., Patterson N.J., Moorjani P. et al.The Genomic Formation of South and Central Asia // Preprint version. – 2018. doi: https://doi.org/10.1101/292581 https://www.biorxiv.org/content/ 10.1101/292581v1
6 Bird S.C. Towards Improvements in the Estimation of the Coalescent: Implications for the Most Effective Use of Y Chromosome Short Tandem Repeat Mutation Rates // PLoS ONE. – 2012. – Vol.7, – N.10. – P.1-11.
7 Purps J., Siegert S., Willuweit S. et al. Global analysis of Y-chromosomal haplotype diversity for 23 STR loci // Forensic Science International: Genetics. – 2014. – Vol.12. – P.12-23.
8 Dabney J., Meyer M., Pääbo S. Ancient DNA damage // Cold Spring Harb. Perspect Biol. – 2013. – Vol.5(7). doi:10.1101/cshperspect.a012567
9 Meyer M. and Kircher M. Illumina sequencing library preparation for highly multiplexed target capture and sequencing // Cold Spring Harb. Protocols. – 2010. – Vol.6. doi: 10.1101/pdb.prot5448.
10 Kircher M., Sawyer S., Meyer M. Double indexing overcomes inaccuracies in multiplex sequencing on the Illumina platform // Nucleic Acids Res. – 2012. – Vol.40(1). doi: 10.1093/nar/gkr771.
11 Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads // EMBnet.journal. – 2011. – Vol.17. P.10-12.
12 Haak W., Balanovsky O., Sanchez J.J. et al. Ancient DNA from European early Neolithic farmers reveals their near eastern affinities // PLoS Biol. – 2010. – Vol.8(11). https://doi.org/10.1371 /journal.pbio.1000536
13 Peltzer A., Jäger G., Herbig A. et al. EAGER: Efficient ancient genome reconstruction // Genome Biol. – 2016. – P.17-60.doi: 10.1186/s13059-016-0918-z.
14 Díaz-Zabala H.J., Nieves-Colón M.A., Martínez-Cruzado J.C. A Mainly Circum-Mediterranean Origin for West Eurasian and North African mtDNAs in Puerto Rico with Strong Contributions from the Canary Islands and West Africa // Human Biology. – 2017. – Vol.89(2). – P.125-155.
15 Underhill P.А., Poznik G.D., Rootsi S. et.al. The phylogenetic and geographic structure of Y-chromosome haplogroup R1a // European Journal of Human Genetics. – 2015. – Vol.23. – P.124-131.
Л.Б. Жансүгірова1, Н. Кағыбатқызы1, З.С. Самашев2, Б.O. Бекманов1
1 ҚР БҒМ ҒК «Жалпы генетика және цитология институты» РМК,Алматы, Қазақстан
2 «Берел» мемлекеттік тарихи-мәдени қорық-мұражайы, Шығыс Қазақстан облысы, Қатонқарағай ауданы, Жамбыл а., Қазақстан
ЕЛЕКЕ САЗЫ ҚОРЫМЫНАН ТАБЫЛҒАН ЕРТЕ КЕЗЕҢГЕ ЖАТАТЫН СҮЙЕК ҚАЛДЫҚТАРЫНЫҢ ГАПЛОТОПТАРЫН АНЫҚТАУ
Аннотация. Елеке Сазы қорымынан табылған (Катон-Қарағай ауданы, Шығыс Қазақстан облысы) ерте темір дәуіріне жататын үш адамның сүйек қалдықтарына палеогенетикалық талдаулар жүргізілді. №4-ші қорғаннан табылған элиталық сақ тұқымына жататын ер адамның Y-хромосомасы бойынша гаплотипі R1a1a1-M417 типімен сипатталды. Сондай-ақ, осы қорғаннан табылған ер және әйел адамдардың мтДНҚ бойынша гаплотоптары J1b1a1e болатыны анықталды. №8-ші қорғаннан табылған элиталық сақ тұқымына жататын егде жастағы әйел адамның гаплотобы A типіне жататыны анықталды.
Түйін сөздер: палеогенетика, ерте кезеңге жататын сақтар, Y-хромосома, мтДНҚ, гаплотоп.
Л.Б. Джансугурова1, Н. Кагыбаткызы1, З.С. Самашев2, Б.O. Бекманов1
1 РГП «Институт общей генетики и цитологии» КН МОН РК,Алматы, Казахстан
2 Государственный историко-культурный музей-заповедник «Берел», Восточно-Казахстанская область,
Катон-Карагайский район, с.Жамбыл, Казахстан
АНАЛИЗ ОТЦОВСКИХ И МАТЕРИНСКИХ ЛИНИЙ ДРЕВНИХ ОБЪЕКТОВ ИЗ НЕКРОПОЛЯ ЕЛЕКЕ САЗЫ
Резюме
Был проведен палеогенетический анализ человеческих останков трех объектов эпохи раннего железного века из некрополя Елеке Сазы (Катон-Карагайский район Восточно-Казахстанская область). Было показано, что отцовская линия элитного сака (курган №4) характеризовалась гаплотипом R1a1a1-M417 Y-хромосомы. J1b1a1e гаплотип мтДНК был определен для мужчины и молодой женщины из №4 кургана. Материнская линия пожилой элитной сакской женщины (№8 курган) принадлежала к A гаплотипу мтДНК.
Ключевые слова: палеогенетика, ранние саки, Y-хромосома, мтДНК, гаплогруппа.



